Overview
As a fundamental component in translation, transfer RNAs (tRNAs) serve as the physical link between the nucleotide sequence of mRNAs and the amino acid sequence of proteins (Fig.1). tRNAs are ubiquitous nucleic acid entities that are the most abundant of all small non-coding RNA molecules. Despite this universality, genomes exhibit substantial variations in their preference for specific codons across their coding sequences. The source of this bias, though still debated, likely reflects selection for translational efficiency and accuracy[1-3].
A wide variety of biological processes, such as cell proliferation[4], differentiation[4, 5] and apoptosis[6], are always accompanied with variation of tRNAs levels. Alterations of tRNA repertoire affect cell-fate choices during cell development (Fig. 2). Many diseases show disruptions to the levels and distributions of tRNAs, such as type 2 diabetes mellitus[19], Huntington disease[16], and HIV infection[18]. Dysregulated tRNA repertoire can promote tumorigenesis and cancer progression[5, 8-15]. tRNA repertoire has become an important aspect in the study of biological processes and human diseases.
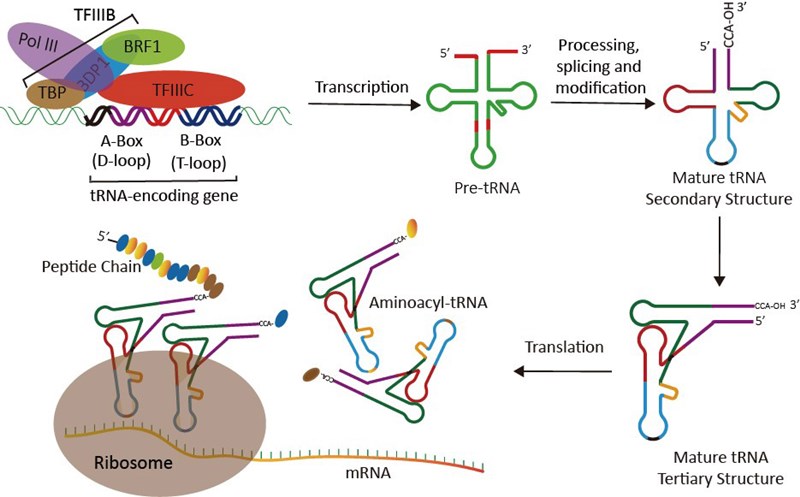
Figure 1. tRNA: the role, function and biogenesis.
tRNA repertoire and its functional significance
Alterations of tRNA levels can profoundly change the cell state by various mechanisms. For example, codon usage is different between the genes serving cell-autonomous functions and the genes involved in multicellularity. tRNAs induced by proliferation and differentiation often carry anticodons that correspond to the codons enriched for these genes accordingly (Fig. 2), which suggests a coordination between tRNA production and mRNA translation[4]. Overexpression of initiator tRNAi(Met) significantly alters the global tRNA expression profile and increases the cell metabolic activity and cell proliferation[5]. At the level of cytochrome c-mediated apoptosome formation, tRNAs can regulate apoptotic sensitivity[6]. Microinjection of tRNA can inhibit cytochrome c-induced apoptosis[7].

Figure 2. tRNA pools are coordinated with the alterations in the mRNA transcriptomes with different codon usage under differentiation or proliferation conditions. The repertoire has the effects on the cell fate determination.
tRNA repertoire and disease
tRNA repertoire has fundamental impact in human diseases. Many diseases are associated with the disrupted tRNAs levels. Dysregulation of certain tRNAs can induce tumorigenesis and cancer progression.
Cancer
After cataloging the tRNA repertoire, Gingold et al demonstrated the tRNA pools are different between cancer and differentiated non-cancer cells[4]. tRNAs that are upregulated in differentiated/arrested cells are repressed in proliferating cells. Conversely, tRNAs whose levels are high in proliferating cells become low in differentiated/arrested cells. Cancer cells adjust their tRNA pools to selectively bolster translation of the mRNAs that are required for tumor progression. By comparing tRNA expression in tumor versus normal breast tissues, Pavon-Eternod et al found that nuclear- and mitochondrial-encoded tRNAs exhibit distinct expression patterns, indicating the potential of using tRNAs as biomarkers for breast cancers[8]. Recently, Goodarzi et al confirmed that specific tRNAs are upregulated in human breast cancer cells as they gain metastatic activity[9]. Further studies showed tRNAGlu-UUC and tRNAArg-CCG promote breast cancer metastasis by directly enhancing EXOSC2 and GRIPAP1 expression. These and other cases conclusively demonstrate dysregulated tRNA repertoire can promote tumorigenesis and cancer progression[5, 8-15].
Huntington’s disease
Huntington disease (HD) is a dominantly inherited neurodegenerative disorder caused by the expansion of a CAG-encoded polyglutamine (polyQ) repeat in huntingtin (Htt). The disease displays a highly heterogeneous etiopathology and disease onset. Analyses of HD-affected brain tissues revealed traces of polyalanine (polyA) or polyserine (polyS) proteins within the polyQ aggregates. These species probably result from a shift in the Gln-encoding CAG frame to an Ala-encoding -1 GCA frame or a Ser-encoding 1 AGC frame. But what is the role of translational frameshifting in the pathogenesis of polyQ diseases? Girstmair et al found that depletion of tRNAGln-CUG pairing to the CAG codon was the main cause of -1 frameshifting. In addition, frameshifted proteins form morphologically distinct aggregates in vivo dependent on the Q:A ratio. The results suggest that frameshifting within expanded CAG stretches may contribute to the heterogeneity in the course and onset of HD on both cellular and individual level[16].
Virus Infection
Viruses are wholly dependent on the host translation machinery to synthesize their proteins. Consequently, viral codon usage is thought to be under selective pressure to adapt to the host cell tRNA pool. Since host codon usage generally reflects the host tRNA pool, viral translation should be more efficient when viral codon usage is similar to that of the host genes. In many cases, however, viral codon usage seems poorly adapted to that of its host. After profiling the tRNA repertoire, Pavon-Eternod et al found that influenza A and vaccinia viruses can manipulate tRNA populations to favor translation of their own genes[17]. HIV-1 is expressed extremely well in human host cells despite their codons are poorly adapted to human host. In another research, it is found that the codon usage of HIV-1 early genes is similar to that of the highly expressed human genes, whereas the codon usage of the late genes is better adapted to the altered tRNA pool induced late in viral infection[18]. This is a striking example of the virus modulating the tRNA pool to optimize its translation efficiency.
tRNA Roadmap
PCR Arrays are the reliable and accurate tools for analyzing the tRNA repertorie. Armed with the tRNA repertoire data, gain-of-function[5, 20, 16] and lose-of-function[17] approaches are useful for follow up studies. Commonly used methods in non-coding RNA studies are readily applicable to in-depth tRNA research.

Figure 3. tRNA research roadmap
Related Products
nrStar™ tRNA PCR Array
nrStar™ tRF&tiRNA PCR Array
Reference
[1] Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 2008;134:341-52.
[2] Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nature reviews Genetics 2011;12:32-42.
[3] Shah P, Gilchrist MA. Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift. Proceedings of the National Academy of Sciences of the United States of America 2011;108:10231-6.
[4] Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 2014;158:1281-92.
[5] Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. Rna 2013;19:461-6.
[6] Mei Y, Stonestrom A, Hou YM, Yang X. Apoptotic regulation and tRNA. Protein & cell 2010;1:795-801.
[7] Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, et al. tRNA binds to cytochrome c and inhibits caspase activation. Molecular cell 2010;37:668-78.
[8] Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic acids research 2009;37:7268-80.
[9] Goodarzi H, Nguyen HC, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell 2016;165:1416-27.
[10] Berns A. A tRNA with oncogenic capacity. Cell 2008;133:29-30.
[11] Waldman YY, Tuller T, Sharan R, Ruppin E. TP53 cancerous mutations exhibit selection for translation efficiency. Cancer research 2009;69:8807-13.
[12] Kushner JP, Boll D, Quagliana J, Dickman S. Elevated methionine-tRNA synthetase activity in human colon cancer. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine 1976;153:273-6.
[13] Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell 2008;133:78-89.
[14] Zhou Y, Goodenbour JM, Godley LA, Wickrema A, Pan T. High levels of tRNA abundance and alteration of tRNA charging by bortezo*** in multiple myeloma. Biochemical and biophysical research communications 2009;385:160-4.
[15] Begley U, Sosa MS, Avivar-Valderas A, Patil A, Endres L, Estrada Y, et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-alpha. EMBO molecular medicine 2013;5:366-83.
[16] Girstmair H, Saffert P, Rode S, Czech A, Holland G, Bannert N, et al. Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin. Cell reports 2013;3:148-59.
[17] Pavon-Eternod M, David A, Dittmar K, Berglund P, Pan T, Bennink JR, et al. Vaccinia and influenza A viruses select rather than adjust tRNAs to optimize translation. Nucleic acids research 2013;41:1914-21.
[18] van Weringh A, Ragonnet-Cronin M, Pranckeviciene E, Pavon-Eternod M, Kleiman L, Xia X. HIV-1 modulates the tRNA pool to improve translation efficiency. Molecular biology and evolution 2011;28:1827-34.
[19] Krokowski D. et al. (2013) "A self-defeating anabolic program leads to beta-cell apoptosis in endoplasmic reticulum stress-induced diabetes via regulation of amino acid flux." J. Biol. Chem. 288(24):17202-13 [PMID: 23645676]
[20] Gong M. et al. (2006) "Overexpression of tnaC of Escherichia coli inhibits growth by depleting tRNA2Pro availability." J. Bacteriol. 188(5):1892-8 [PMID: 16484200]
[21] Yona A.H. et al. (2013) "tRNA genes rapidly change in evolution to meet novel translational demands." Elife 2:e01339 [PMID: 24363105]
[22] Fu G. et al. (2012) "tRNA-controlled nuclear import of a human tRNA synthetase." J. Biol. Chem. 287(12):9330-4 [PMID: 22291016]