Related Products
Related Services
Related Reviews
G4 CUT&Tag Library Prep Kit
Arraystar G4 CUT&Tag Library Prep Kit is designed for constructing G4 CUT&Tag sequencing library to map and profile DNA quadruplexes in the chromatin genome-wide. For the constructed libraries, Arraystar offers sequencing and bioinformatics analysis service.
Benefits:
• High resolution, specificity, sensitivity, and reproducibility: High-resolution mapping of G4 sites within a few bases. Highly specific and sensitive antibody targeting for superior G4s signals. The results are highly reproducible across technical replicates.
• G4 detection in enhancer, promoter, and gene body regions, even for transient G4s in vivo.
• Convenience: Tagmentation is much easier to perform than ChIP-seq based methods. No optimization of fixation, sonication, digestion, or immunoprecipitation required.
• Improved sample prep: Cells are collected by centrifugation instead of original ConA magnetic beads, which is applicable to more sample types (e.g. most cell types or frozen tissues) at higher cell viability (> 85%).
| Product Name | Catalog No. | Description | Size | Price |
|---|---|---|---|---|
| G4 CUT&Tag Library Prep Kit | AS-TN-006 | 12 reactions | $1,198.00 |
G-quadruplex (G4) is a non-canonical nucleic acid structure that arises from the self-stacking of two or more G-quartets formed by Hoogsteen hydrogen bonds between the guanines. G-quadruplexes in DNA (dG4) are distributed in specific genomic regions and implicated in several essential cellular processes.
Arraystar G4 CUT&Tag Library Prep Kit is designed for constructing G4 CUT&Tag sequencing library to map and profile DNA quadruplexes in the chromatin genome-wide. In the CUT&Tag (Cleavage Under Targets & Tagmentation), BG4 antibody is used to bind G4s. The antibody bound G4 sites are then fragmented and transposon tagged by protein A-Tn5 transposase fusion enzyme in a single tagmentation step. This method is rapidly replacing traditional chromatin immunoprecipitation sequencing (ChIP-seq) due to its low background, high signal-to-noise ratio, better reproducibility, and ease of use.
As an example, the G4 CUT&Tag library was prepared using 200,000 Hela cells with the kit according to the kit protocol. The sequencing results are displayed in Fig. 1 and 2.
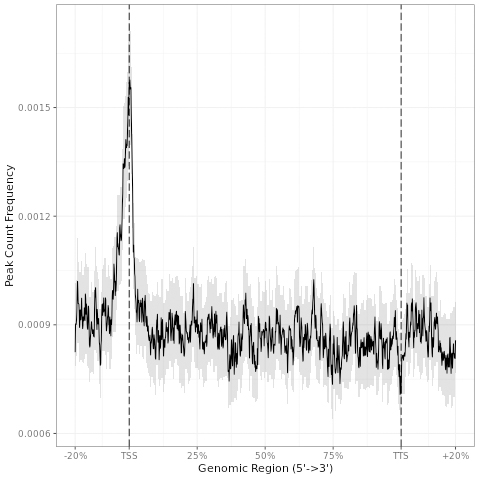
Fig.1. The G4 peak distribution in standardized transcription unit, showing significant enrichment in promoter region (TSS).

Fig.2. An IGV view of G4 signals by G4-ChIP (upper track, high noise) and G4-CUT&Tag (lower track, low noise).

