Arraystar R-loop Profiling Service profiles LncRNA/mRNA organized R-loop distribution in the genome. S9.6 antibody is used to the highly specifically immunoprecipitate the R-loops. The R-loop Profiling data are bioinformatically analyzed to gain biological and functional insights.
Along with LncRNA Array, MeDIP-seq, or ChIP-seq, R-loop Profiling Service provides valuable functional insights in epigenetic and transcriptional regulation by R-loops.
Benefits
• Powerful profiling: To study R-loops as a new player in gene regulation in the genome.
• High reliability: Well established optimal experimental procedures to produce best possible results.
• Flexibility: The R-loop Profiling Service can be performed for any species with a reference genome.
• Rigorous quality: The positive and negative controls ensure library quality.
• Rich annotation: RNA type (lncRNA or mRNA) in R-loop, R-loop peak GC skew values, classification, distribution & enrichment in gene features, and distribution within gene body
• Visualization: Provided with genome browser tracks, and publication-ready graphics.
Watch Video> R-Loops: An Active Player in Cancer and Diseases
R-loop is a RNA:DNA three-stranded hybrid structure formed between LncRNA/mRNA/CircRNA strand and the template DNA strand by base pairing, leaving the non-template DNA strand unpaired and displaced in the loop (Fig.1) [1]. R-loops are widely distributed, occurring in 5% of the mammalian genomes [2.3]. R-loops are often located in the CpG islands of the promoters or transcription stop sites. High GC skews (G enrichment over C in the downstream of TSS non-template strand), G-quadruplexes, DNA gaps and DNA/RNA modifications contribute the formation of R-loops [4]. R-loops have important biological functions in gene regulation, DNA replication, and DNA/histone modifications.

Figure 1. R-loop structure[5].
Antisense lncRNA organized R-loops impact mRNA transcription
R-loops can form between an antisense RNA strand and the DNA in the loop. TCF21 is a tumor suppressor in many cancers. TARID (TCF21 antisense RNA inducing demethylation) is a head-to-head antisense lncRNA of TCF21 gene and forms R-loop in the promoter region (Fig. 3). The R-loop is recognized by GADD45a, which recruits demethylase TET1, removes DNA methylation, increases TCF21 mRNA transcription, and regulates the cell cycles [7].
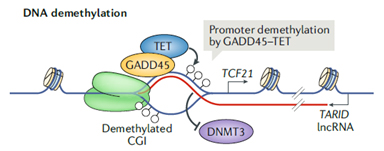
Figure 3. Antisense lncRNATARID forms R-loop to regulate TCF21 promoter demethylation and TCF21 mRNA transcription [4].
mRNA organized R-loop regulation of DNA methylation and mRNA transcription
Normally, the R-loop at BAMBI (a negative regulator of TGFb) gene promoter facilitates more transcription. However in amyotrophic lateral sclerosis (ALS4), senataxin mutation reduces R-loop and increases the DNA methylation at the BAMBI promoter, leading to BAMBI transcription repression, TGFb signal transduction upregulation, and ALS progression (Fig. 2)[6].

Figure 2. In healthy normal cell, R-loop at the BAMBI promoter facilitates more transcription. In ALS cells, Senataxin mutation suppresses R-loop formation at the promoter and BAMBI transcription.
Related Review
Why R-loop is an active player in transcription regulation?
References
[1] Hamperl S. and Cimprich KA. (2014) The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst) 19:84-94 [PMID:24746923]
[2] Sanz LA. et al (2016) Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell 63(1):167-78 [PMID:27373332]
[3] Li M. and Klungland A. (2020) Modifications and interactions at the R-loop. DNA Repair (Amst) 96:102958 [PMID:32961406]
[4] Niehrs C. and Luke B. (2020) Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol 21(3):167-178 [PMID:32005969]
[5] Hegazy YA. et al (2020) The balancing act of R-loop biology: The good, the bad, and the ugly. J Biol Chem 295(4):905-913 [PMID:31843970]
[6] Grunseich C. et al (2018) Senataxin Mutation Reveals How R-Loops Promote Transcription by Blocking DNA Methylation at Gene Promoters. Mol Cell 69(3):426-437.e7 [PMID:29395064]
[7] Arab K. et al (2019) GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat Genet 51(2):217-223 [PMID:30617255]

Fig 1. The workflow of R-loop Profiling Service